Pandora: 4-D White Matter Bundle Population-Based Atlases Derived from Diffusion MRI Fiber Tractography
Colin B. Hansen, Qi Yang, Ilwoo Lyu, Francois Rheault, Cailey Kerley, Bramsh Qamar Chandio, Shreyas Fadnavis, Owen Williams, Andrea T. Shafer, Susan M. Resnick, David H. Zald, Laurie E. Cutting, Warren D. Taylor, Brian Boyd, Eleftherios Garyfallidis, Adam W. Anderson, Maxime Descoteaux, Bennett A. Landman, Kurt G. Schilling. Pandora: 4-D White Matter Bundle Population-Based Atlases Derived from Diffusion MRI Fiber Tractography. Neuroinform (2020).
Abstract
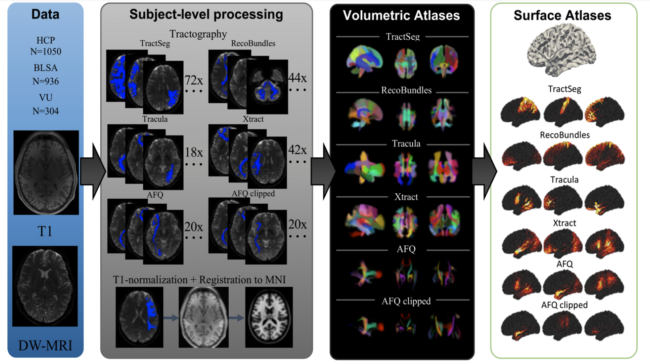
Brain atlases have proven to be valuable neuroscience tools for localizing regions of interest and performing statistical inferences on populations. Although many human brain atlases exist, most do not contain information about white matter structures, often neglecting them completely or labelling all white matter as a single homogenous substrate. While few white matter atlases do exist based on diffusion MRI fiber tractography, they are often limited to descriptions of white matter as spatially separate “regions” rather than as white matter “bundles” or fascicles, which are well-known to overlap throughout the brain. Additional limitations include small sample sizes, few white matter pathways, and the use of outdated diffusion models and techniques. Here, we present a new population-based collection of white matter atlases represented in both volumetric and surface coordinates in a standard space. These atlases are based on 2443 subjects, and include 216 white matter bundles derived from 6 different automated state-of-the-art tractography techniques. This atlas is freely available and will be a useful resource for parcellation and segmentation.