Semi-supervised Machine Learning with MixMatch and Equivalence Classes
Colin B. Hansen, Vishwesh Nath, Riqiang Gao, Camilo Bermudez, Yuankai Huo, Kim L. Sandler, Pierre P. Massion, Jeffrey D. Blume, Thomas A. Lasko, Bennett A. Landman “Semi-supervised Machine Learning with MixMatch and Equivalence Classes.” Interpretable and Annotation-Efficient Learning for Medical Image Computing. Springer, Cham, 2020. 112-121.
Abstract
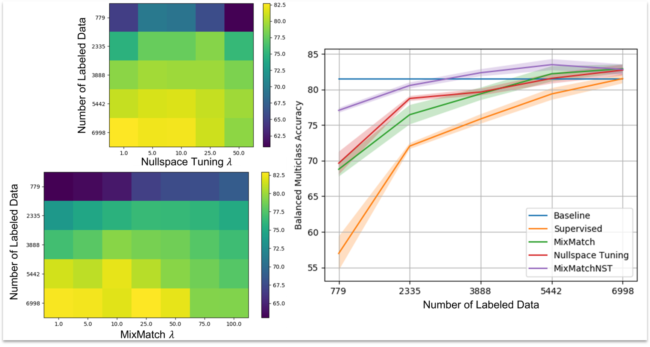
Semi-supervised methods have an increasing impact on computer vision tasks to make use of scarce labels on large datasets, yet these approaches have not been well translated to medical imaging. Of particular interest, the MixMatch method achieves significant performance improvement over popular semi-supervised learning methods with scarce labels in the CIFAR-10 dataset. In a complementary approach, Nullspace Tuning on equivalence classes offers the potential to leverage multiple subject scans when the ground truth for the subject is unknown. This work is the first to (1) explore MixMatch with Nullspace Tuning in the context of medical imaging and (2) characterize the impacts of the methods with diminishing labels. We consider two distinct medical imaging domains: skin lesion diagnosis and lung cancer prediction. In both cases we evaluate models trained with diminishing labeled data using supervised, MixMatch, and Nullspace Tuning methods as well as MixMatch with Nullspace Tuning together. MixMatch with Nullspace Tuning together is able to achieve an AUC of 0.755 in lung cancer diagnosis with only 200 labeled subjects on the National Lung Screening Trial and a balanced multi-class accuracy of 77% with only 779 labeled examples on HAM10000. This performance is similar to that of the fully supervised methods when all labels are available. In advancing data driven methods in medical imaging, it is important to consider the use of current state-of-the-art semi-supervised learning methods from the greater machine learning community and their impact on the limitations of data acquisition and annotation.
Keywords: Semi-supervised learning, skin lesion, lung cancer