Generalizing deep whole-brain segmentation for post-contrast MRI with transfer learning
Abstract
Purpose: Generalizability is an important problem in deep neural networks, especially with variability of data acquisition in clinical magnetic resonance imaging (MRI). Recently, the spatially localized atlas network tiles (SLANT) can effectively segment whole brain, non-contrast T1w MRI with 132 volumetric labels. Transfer learning (TL) is a commonly used domain adaptation tool to update the neural network weights for local factors, yet risks degradation of performance on the original validation/test cohorts.
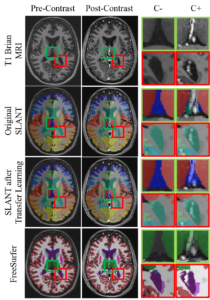
Approach: We explore TL using unlabeled clinical data to address these concerns in the context of adapting SLANT to scanning protocol variations. We optimize whole-brain segmentation on heterogeneous clinical data by leveraging 480 unlabeled pairs of clinically acquired T1w MRI with and without intravenous contrast. We use labels generated on the pre-contrast image to train on the post-contrast image in a five-fold cross-validation framework. We further validated on a withheld test set of 29 paired scans over a different acquisition domain.
Results: Using TL, we improve reproducibility across imaging pairs measured by the reproducibility Dice coefficient (rDSC) between the pre- and post-contrast image. We showed an increase over the original SLANT algorithm (rDSC 0.82 versus 0.72) and the FreeSurfer v6.0.1 segmentation pipeline (rDSC = 0.53). We demonstrate the impact of this work decreasing the root-mean-squared error of volumetric estimates of the hippocampus between paired images of the same subject by 67%.
Conclusion: This work demonstrates a pipeline for unlabeled clinical data to translate algorithms optimized for research data to generalize toward heterogeneous clinical acquisitions.