High-resolution 3D abdominal segmentation with random patch network fusion
Y. Tang,R.Gao,S.Han, Y.Chen, D.Gao, V.Nath, C.Bermudez, M.R. Savona, R.G. Abramson, S.Bao,I.Lyu, Y.Huo and B.A. Landman,“High-resolution 3D Abdominal Segmentation with Random PatchNetworkFusion”,Medical Image Analysis, 2021.
Full Text:
https://www.sciencedirect.com/science/article/pii/S1361841520302589
Abstract
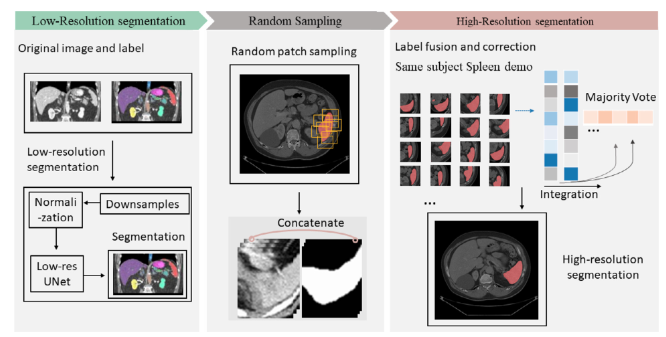
Deep learning for three dimensional (3D) abdominal organ segmentation on high-resolution computed to- mography (CT) is a challenging topic, in part due to the limited memory provide by graphics processing units (GPU) and large number of parameters and in 3D fully convolutional networks (FCN). Two preva- lent strategies, lower resolution with wider field of view and higher resolution with limited field of view, have been explored but have been presented with varying degrees of success. In this paper, we propose a novel patch-based network with random spatial initialization and statistical fusion on overlapping regions of interest (ROIs). We evaluate the proposed approach using three datasets consisting of 260 subjects with varying numbers of manual labels. Compared with the canonical “coarse-to-fine” baseline methods, the proposed method increases the performance on multi-organ segmentation from 0.799 to 0.856 in terms of mean DSC score (p-value < 0.01 with paired t -test). The effect of different numbers of patches is eval- uated by increasing the depth of coverage (expected number of patches evaluated per voxel). In addition, our method outperforms other state-of-the-art methods in abdominal organ segmentation. In conclusion, the approach provides a memory-conservative framework to enable 3D segmentation on high-resolution CT. The approach is compatible with many base network structures, without substantially increasing the complexity during inference. Given a CT scan with at high resolution, a low-res section (left panel) is trained with multi-channel seg- mentation. The low-res part contains down-sampling and normalization in order to preserve the complete spatial information. Interpolation and random patch sampling (mid panel) is employed to collect patches. The high-dimensional probability maps are acquired (right panel) from integration of all patches on field of views.